-
Notifications
You must be signed in to change notification settings - Fork 283
Description
I have been using reconstruction software written in IDL at APS beamline 13-BM-D. It uses gridrec (tomoRecon) written in C++ at its core, but uses IDL as the user interface and programming language.
I want to switch to using tomopy. I am comparing results and performance between IDL and tomopy, and I am having problems with entropy based centering. The problem may just be that I don't understand how to use it in tomopy.
For IDL I first did the centering with the following configuration:
• Sinogram padding to 2048 (X size of projections is 1920)
• 10 pixel average of air pixels
• No ring artifact removal
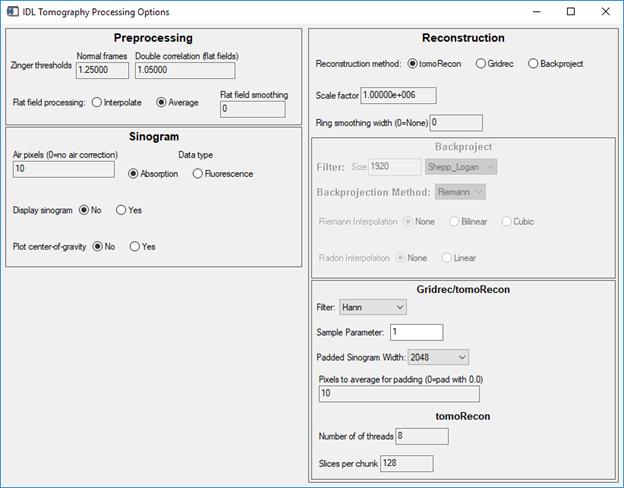
This is a screen shot of the configuration screen. Note that Air pixels=10, Ring smoothing width=0, Padded Sinogram Width=2048.
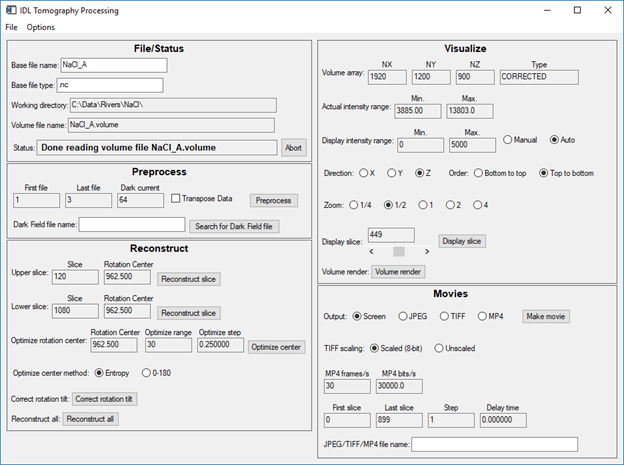
This is a screen shot of the main IDL screen. In the Reconstruct section I have set Optimize method=Entropy, the initial guess of the rotation center=960, Optimize range=30 (+- 15 pixels), and the Optimize step=0.25 pixels. This will reconstruct the upper and lower slices at 120 different centers (30 pixels at 0.25 pixel step). I have selected an upper slice=120 and a lower slice=1080 to do the centering.
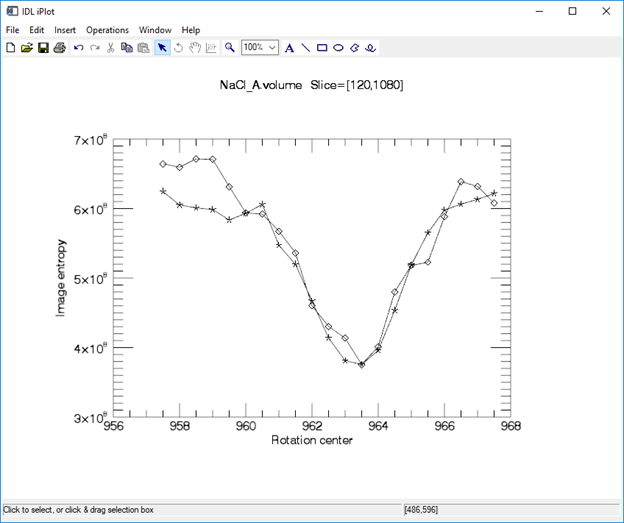
When I press optimize center I get this plot of the entropy as a function of rotation center. One plot is for the upper slice, the other for the lower slice. These curves show that the entropy is an extremely smooth function of rotation center, with single very well-defined minimum.
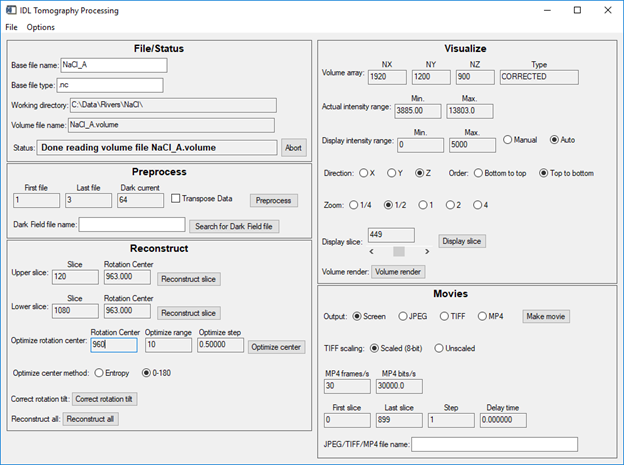
This is the main screen after optimization. Notice that the rotation center for both the upper and lower slices is 962.50. Thus, within the step size of 0.25 pixels the rotation center is the same for slices 120 and 1080. This means that the rotation axis is very well aligned with the columns of the camera.
I changed my initial guess of the center from 960 to 970. That resulted in this plot.
The minima were found in the identical location, 962.5.
I repeated the optimization again, but starting with an initial guess of 950. That also resulted in exactly the same center for both slices, 962.5
I optimized over a range of 30 pixels for this example. That required 26.7 seconds total to optimize both slices. In practice it is usually sufficient to optimize over a range of 10 pixels, since the rotation axis typically does not change during an experiment by more than +-5 pixels. Optimizing over a 10 pixel range takes 8.9 seconds.
In IDL I can also find the center using the 0 and 180 views and subtracting the images.
This is the setup for that. Note that Optimize center method=0-180 and the Optimize step is 0.5, which is the maximum resolution for this method.
This is the resulting plot. Note that the Y axis label is incorrect, it is not entropy it is the difference between the 0 and flipped 180 image as a function of the shift in one image.
This technique found the center at 963.0, within the 0.5 pixel step size of the value found by the entropy method.
Optimizing the center using tomopy
I used this Python program to test finding the centers with tomopy. It is intended to do the same steps as IDL above.
- Normalize to dark and flat fields
- Normalize to 10 pixels of air
- Pad to 2048 pixel width
- Find the center using the 0-180 method
- Find the center using the entropy method with initial guesses of 960, 950, and 970.
import tomopy
import dxchange
import logging
top_slice=120
bottom_slice = 1080
logging.basicConfig(
format='%(asctime)s %(levelname)-8s %(message)s',
level=logging.INFO,
datefmt='%Y-%m-%d %H:%M:%S')
logging.info('Reading data ...')
proj, flat, dark, theta = dxchange.read_aps_13bm('NaCl_A1.nc', file_format='netcdf4')
logging.info('Normalizing ...')
norm = tomopy.normalize(proj, flat, dark)
logging.info('Secondary normalize to air ...')
norm = tomopy.normalize_bg(norm, air=10)
npad = int((2048 - norm.shape[2]) / 2)
logging.info('Padding to 2048, npad=%d', npad)
norm = tomopy.misc.morph.pad(norm, axis=2, npad=npad, mode='edge')
logging.info('Shape after padding %s', norm.shape)
logging.info('Taking log ...')
norm = tomopy.minus_log(norm)
center_guess = 960 + npad
logging.info("")
logging.info('Finding centers with 0-180')
top_center = tomopy.find_center_pc(norm[0, top_slice:top_slice+10, :], norm[-1, top_slice:top_slice+10, :], rotc_guess=center_guess, tol=0.5)
bottom_center = tomopy.find_center_pc(norm[0, bottom_slice:bottom_slice+10, :], norm[-1, bottom_slice:bottom_slice+10 :], rotc_guess=center_guess, tol=0.5)
logging.info('top_center: %f, bottom_center:%f', top_center-npad, bottom_center-npad)
center_guess = 960 + npad
logging.info("")
logging.info('Finding centers with entropy, initial guess=%f', center_guess)
top_center = tomopy.find_center(norm, theta, init=center_guess, ind=top_slice, tol=0.5)
bottom_center = tomopy.find_center(norm, theta, init=center_guess, ind=bottom_slice, tol=0.5)
logging.info('top_center: %f, bottom_center:%f', top_center, bottom_center)
center_guess = 950 + npad
logging.info("")
logging.info('Finding centers with entropy, initial guess=%f', center_guess)
top_center = tomopy.find_center(norm, theta, init=center_guess, ind=top_slice, tol=0.5)
bottom_center = tomopy.find_center(norm, theta, init=center_guess, ind=bottom_slice, tol=0.5)
logging.info('top_center: %f, bottom_center:%f', top_center, bottom_center)
center_guess = 970 + npad
logging.info("")
logging.info('Finding centers with entropy, initial guess=%f', center_guess)
top_center = tomopy.find_center(norm, theta, init=center_guess, ind=top_slice, tol=0.5)
bottom_center = tomopy.find_center(norm, theta, init=center_guess, ind=bottom_slice, tol=0.5)
logging.info('top_center: %f, bottom_center:%f', top_center, bottom_center)
#logging.info('Reconstructing')
#recon = tomopy.recon(norm, theta, center=top_center, algorithm='gridrec', sinogram_order=False)
#logging.info('Reconstruction complete')I have attached the program here as a .txt file.
test_center.py.txt
This is the output when running that program on the same dataset processed with IDL above. It's dimensions are NX=1920, NY=1200, NProjections=900. I have deleted some of the logging.info output from tomopy.find_center() to save space.
corvette:~/scratch/tomo_data/NaCl>python test_center.py
2020-05-12 15:18:23 INFO Reading data ...
2020-05-12 15:18:30 INFO Data successfully imported: /home/epics/scratch/tomo_data/NaCl/NaCl_A2.nc
2020-05-12 15:18:30 INFO Data successfully imported: /home/epics/scratch/tomo_data/NaCl/NaCl_A1.nc
2020-05-12 15:18:30 INFO Data successfully imported: /home/epics/scratch/tomo_data/NaCl/NaCl_A3.nc
2020-05-12 15:18:31 INFO Normalizing ...
2020-05-12 15:18:36 INFO Secondary normalize to air ...
2020-05-12 15:18:46 INFO Padding to 2048, npad=64
/home/epics/anaconda3/lib/python3.7/site-packages/tomopy/misc/morph.py:141: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
ne.evaluate("arr", out=out[slc_in])
/home/epics/anaconda3/lib/python3.7/site-packages/tomopy/misc/morph.py:148: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
ne.evaluate("vec", local_dict={'vec': arr[slc_l_v]},
/home/epics/anaconda3/lib/python3.7/site-packages/tomopy/misc/morph.py:149: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
out=out[slc_l])
/home/epics/anaconda3/lib/python3.7/site-packages/tomopy/misc/morph.py:150: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
ne.evaluate("vec", local_dict={'vec': arr[slc_r_v]},
/home/epics/anaconda3/lib/python3.7/site-packages/tomopy/misc/morph.py:151: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
out=out[slc_r])
2020-05-12 15:18:48 INFO Shape after padding (900, 1200, 2048)
2020-05-12 15:18:48 INFO Taking log ...
2020-05-12 15:18:49 INFO
2020-05-12 15:18:49 INFO Finding centers with 0-180
2020-05-12 15:18:49 INFO top_center: 962.250000, bottom_center:962.250000
2020-05-12 15:18:49 INFO
2020-05-12 15:18:49 INFO Finding centers with entropy, initial guess=1024.000000
2020-05-12 15:18:49 INFO Trying rotation center: [1024.]
2020-05-12 15:18:50 INFO Function value = 2.493676
2020-05-12 15:18:50 INFO Trying rotation center: [1075.2]
…
2020-05-12 15:19:07 INFO Trying rotation center: [941.2]
2020-05-12 15:19:08 INFO Function value = 2.444963
2020-05-12 15:19:08 INFO top_center: 1076.800000, bottom_center:941.600000
2020-05-12 15:19:08 INFO
2020-05-12 15:19:08 INFO Finding centers with entropy, initial guess=1014.000000
2020-05-12 15:19:08 INFO Trying rotation center: [1014.]
2020-05-12 15:19:08 INFO Function value = 2.494112
2020-05-12 15:19:08 INFO Trying rotation center: [1064.7]
…
2020-05-12 15:19:24 INFO Trying rotation center: [958.94296875]
2020-05-12 15:19:24 INFO Function value = 2.444797
2020-05-12 15:19:24 INFO top_center: 1082.128125, bottom_center:959.339063
2020-05-12 15:19:24 INFO
2020-05-12 15:19:24 INFO Finding centers with entropy, initial guess=1034.000000
2020-05-12 15:19:24 INFO Trying rotation center: [1034.]
2020-05-12 15:19:25 INFO Function value = 2.493973
…
2020-05-12 15:19:39 INFO Trying rotation center: [956.04609375]
2020-05-12 15:19:40 INFO Function value = 2.444716
2020-05-12 15:19:40 INFO top_center: 1072.775000, bottom_center:956.450000m.
Conclusions
Using the 0-180 algorithm in tomopy gives the top and bottom centers as 962.25, which is within 0.25 pixels of the values obtained with the entropy method in IDL (962.5).
However, the following table shows that the centers determined with the entropy method in tomopy are highly scattered, and depend greatly on the initial guess.
| Program | Initial guess | Top slice center | Bottom slice center |
|---|---|---|---|
| IDL | 960 | 962.5 | 962.5 |
| IDL | 950 | 962.5 | 962.5 |
| IDL | 970 | 962.5 | 962.5 |
| Tomopy | 960 | 1076.8 | 941.6 |
| Tomopy | 950 | 1082.1 | 959.3 |
| Tomopy | 970 | 1072.7 | 956.4 |
Am I doing something wrong? Why are the tomopy centers using entropy so wrong, and highly dependent on the initial guess?